|
|
||||||||||||||||||
Dynamic Influence Networks for Rule-based Models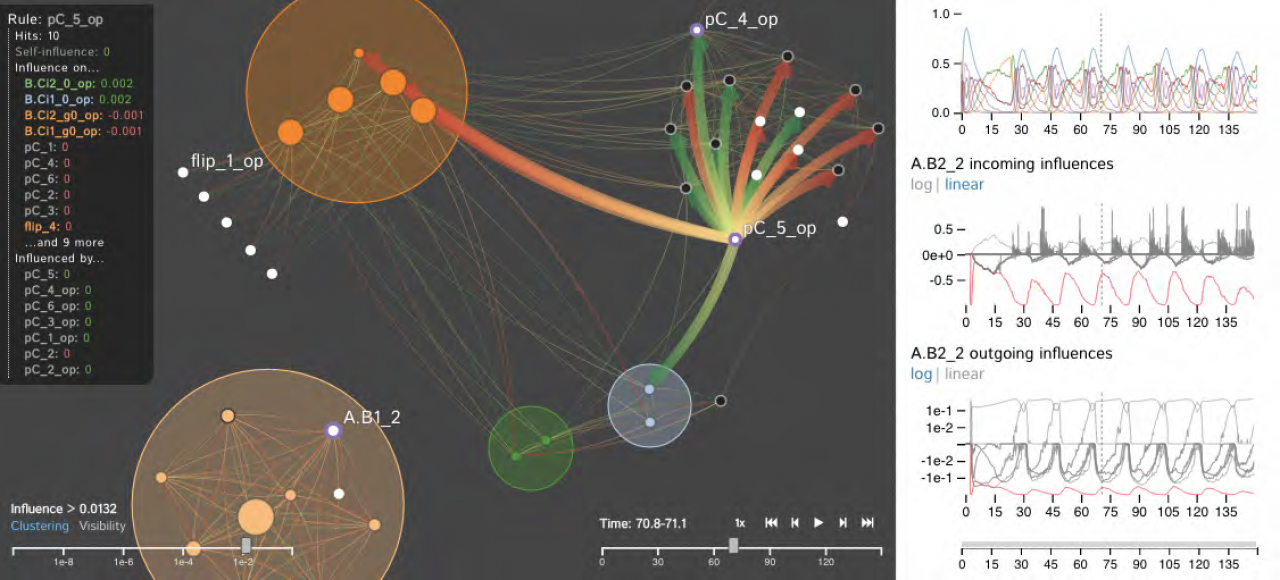
Authors: Forbes, A.G., Burks, A., Lee, K., Li, X., Boutillier, P., Krivine, J., Fontana, W.
Publication: IEEE Transactions on Visualization and Computer Graphics, pp. 1-1 URL: http://dx.doi.org/10.1109/TVCG.2017.2745280 We introduce the Dynamic Influence Network (DIN), a novel visual analytics technique for representing and analyzing rule-based models of protein-protein interaction networks. Rule-based modeling has proved instrumental in developing biological models that are concise, comprehensible, easily extensible, and that mitigate the combinatorial complexity of multi-state and multi-component biological molecules. Our technique visualizes the dynamics of these rules as they evolve over time. Using the data produced by KaSim, an open source stochastic simulator of rule-based models written in the Kappa language, DINs provide a node-link diagram that represents the influence that each rule has on the other rules. That is, rather than representing individual biological components or types, we instead represent the rules about them (as nodes) and the current influence of these rules (as links). Using our interactive DIN-Viz software tool, researchers are able to query this dynamic network to find meaningful patterns about biological processes, and to identify salient aspects of complex rule-based models. To evaluate the effectiveness of our approach, we investigate a simulation of a circadian clock model that illustrates the oscillatory behavior of the KaiC protein phosphorylation cycle. Index Terms - Dynamic networks, biological data visualization, rule-based modeling, protein-protein interaction networks. Date: August 1, 2017 Document: View PDF |