|
|
||||||||||||||||||
Explainable Spatial Clustering: Leveraging Spatial Data in Radiation Oncology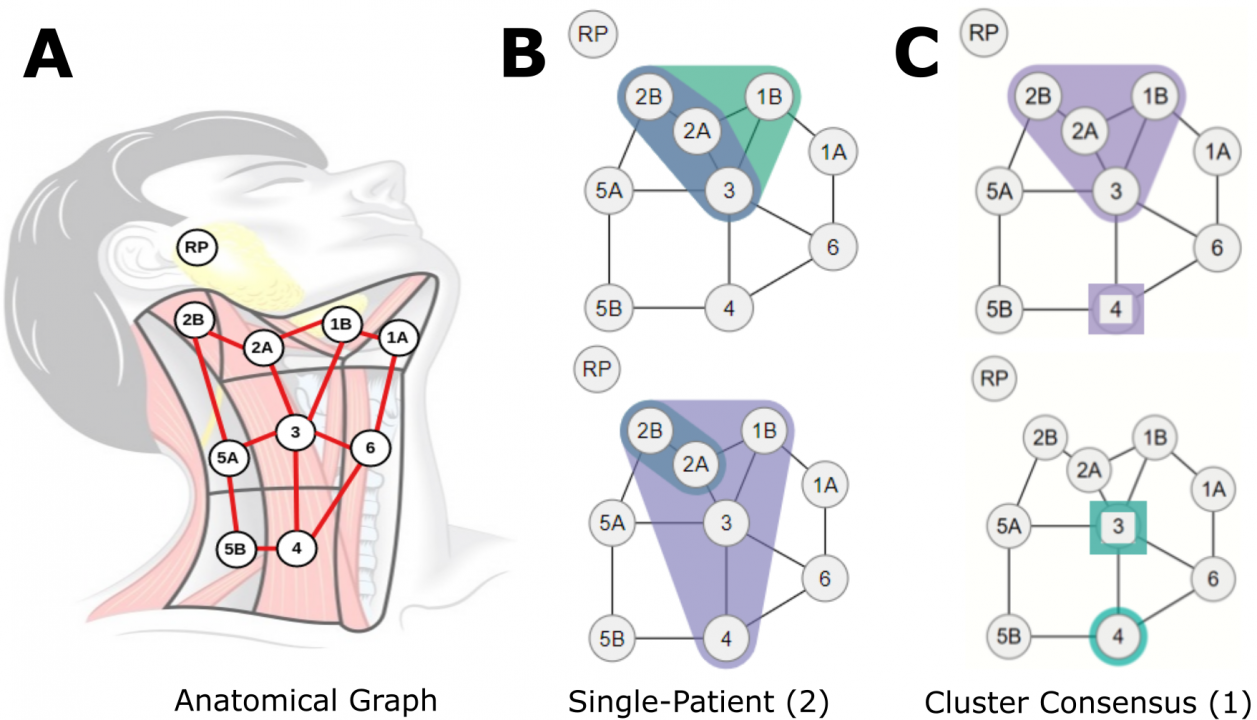
Authors: Wentzel, A., Canahuate, G., van Dijk, L., Mohamed, A., Fuller, C.D., Marai, G.E.
Publication: IEEE VIS 2020 Short Papers, Visualizing Machine Learning URL: https://virtual.ieeevis.org/paper_s-short-1207.html Advances in data collection in radiation therapy have led to an abundance of opportunities for applying data mining and machine learning techniques to promote new data-driven insights. In light of these advances, supporting collaboration between machine learning experts and clinicians is important for facilitating better development and adoption of these models. Although many medical use-cases rely on spatial data, where understanding and visualizing the underlying structure of the data is important, little is known about the interpretability of spatial clustering results by clinical audiences. In this work, we reflect on the design of visualizations for explaining novel approaches to clustering complex anatomical data from head and neck cancer patients. These visualizations were developed, through participatory design, for clinical audiences during a multi-year collaboration with radiation oncologists and statisticians. We distill this collaboration into a set of lessons learned for creating visual and explainable spatial clustering for clinical users. Keywords: Data Clustering and Aggregation, Life Sciences, Collaboration, Mixed Initiative Human-Machine Analysis, Guidelines YouTube Video: https://youtu.be/UM8-_MN3a2Y Date: October 25, 2020 - October 30, 2020 Document: View PDF |