|
|
||||||||||||||||||
Oropharyngeal cancer patient stratification using random forest based‑learning over high‑dimensional radiomic features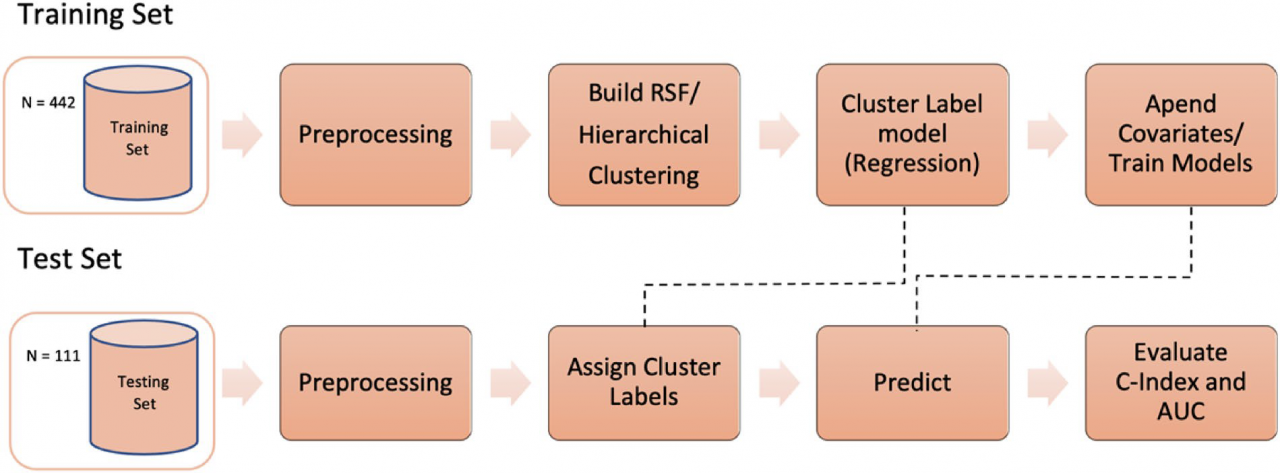
Authors: Patel, H., Vock, D., Marai, G.E., Fuller, C.D., Mohamed, A.S.R., Canahuate, G.
Publication: Scientific Reports 2021, vol 11 URL: https://doi.org/10.1038/s41598-021-92072-8 To improve risk prediction for oropharyngeal cancer (OPC) patients using cluster analysis on the radiomic features extracted from pre‑treatment Computed Tomography (CT) scans. 553 OPC Patients randomly split into training (80%) and validation (20%), were classified into 2 or 3 risk groups by applying hierarchical clustering over the co‑occurrence matrix obtained from a random survival forest (RSF) trained over 301 radiomic features. The cluster label was included together with other clinical data to train an ensemble model using five predictive models (Cox, random forest, RSF, logistic regression, and logistic‑elastic net). Ensemble performance was evaluated over the independent test set for both recurrence-free survival (RFS) and overall survival (OS). The Kaplan–Meier curves for OS stratified by cluster label show significant differences for both training and testing (p val < 0.0001). When compared to the models trained using clinical data only, the inclusion of the cluster label improves AUC test performance from .62 to .79 and from .66 to .80 for OS and RFS, respectively. The extraction of a single feature, namely a cluster label, to represent the high‑dimensional radiomic feature space reduces the dimensionality and sparsity of the data. Moreover, inclusion of the cluster label improves model performance compared to clinical data only and offers comparable performance to the models including raw radiomic features. Date: July 7, 2021 Document: View PDF |